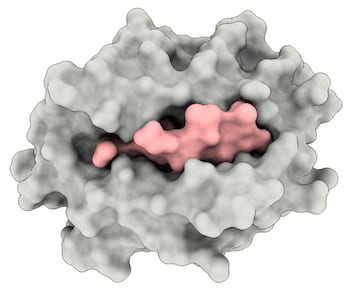
Rice, MD Anderson team models complex that immune system uses to recognize viruses.
“Position 4” didn’t seem important until researchers took a long look at a particular peptide.
That part of the peptide drawn from a SARS-CoV virus turned out to have an unexpected but significant influence on how it stably binds with a receptor central to the immune system’s ability to attack diseased cells.
In a study published by the Proceedings of the National Academy of Sciences, researchers at Rice University’s Brown School of Engineering and the University of Texas MD Anderson Cancer Center revealed models at atomic resolution that detail not only the binding but also, for the first time, the unbinding mechanisms that underlie a key component of the immune system.
They say better understanding of the entire mechanism could lead to advancements in immunotherapy that boosts the body’s ability to fight disease.
Rice computer scientist Lydia Kavraki, alumnus Jayvee Abella and postdoctoral researcher Dinler Antunes, led the study.
“Finding good targets to trigger a protective immune response is very challenging, especially in cancer research,” Antunes said. “The fact that this particular peptide was predicted not to bind to HLAs (human leukocyte antigens) by sequence-based methods highlights a blind spot in our current prediction capacity.
“By incorporating structural analysis, we can detect the contribution of these secondary interactions to peptide binding and stability, hopefully enabling us to find better targets for antiviral vaccine development and T-cell-based cancer immunotherapy,” he said.
The researchers used their simulations to illuminate details of how the intracellular SARS peptide, QFKDNVILL, binds to an MHC receptor protein known as HLA-A∗24:02, primarily at dominant anchors on both ends of the peptide (at positions 2 and 9) and presents them for inspection to the immune system’s T cells.
Stable binding of a peptide and MHC is a prerequisite to the activation of T cells, which look for peptides not normally found in healthy cells. If the peptide and protein don’t bind, the T cell is not prompted to attack.
“That much was known from previous studies of the bound and unbound states of many such complexes,” Kavrakisaid. “What they didn’t capture was the intermediate states and the transitions that lead from one state to another, especially the unbinding.
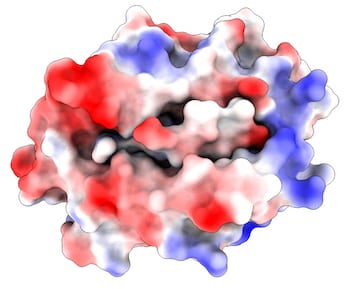
“I think this is the only analysis that shows the unbinding of peptides from the MHC with atomic resolution,” Kavraki said. “Other peptides have similar characteristics and we think they would have similar behaviors.”
All of these interactions were revealed in great detail through Markov state models that analyze how systems change over time. In this case, the models revealed the importance of secondary sites that support the peptide’s primary anchors. That’s where position 4 stood out.
“There are the main, canonical anchors that people know, but there are these secondary interactions that contribute to the binding and the stability,” Antunes said. “These are harder to capture, but in this study, it seems that position 4 plays a very important role. When you mutate it, it affects the behavior of the peptide as it unbinds from the molecule.”
The researchers modeled mutations of the MHC to see how they would influence binding and found they supported the importance of position 4 to the stability of the complex.
“Our computational approach was able to make predictions on the effect of mutations that are then experimentally verified,” said co-author Cecilia Clementi, a former Rice professor who recently became Einstein Professor of Physics at the Free University of Berlin.


The researchers developed a two-stage process to simplify the computational complexity of atom-scale analysis of large molecules. The first stage used a technique called umbrella sampling to accelerate the initial exploration of the molecules. The second, exploratory stage used adaptive sampling, in which simulations are driven to accelerate the construction of the Markov model.
“The challenge is that these MHCs are pretty large systems for computational chemists to simulate,” said Abella, whose research on the topic formed much of his doctoral thesis. “We had to make some approximations and leverage advances in these classes of methods to move forward.
“We’re not the first one to study unbinding, but what characterizes our work over others is that we keep full atomic resolution in our simulations,” he said. “Other works use a technique known as a Markov chain Monte Carlo, whereas we use molecular dynamics, which lets us incorporate time into our computation to capture the kinetics.”
Their methods can be applied to other peptide-MHC complexes with existing 3D models. “This was, in some sense, a feasibility study to show we can use molecular dynamics and build a Markov state model of a system this size,” Abella said.
The researchers also noted the study’s relevance to the current fight against COVID-19, as the SARS peptide they viewed, QFKDNVILL, is highly similar to the NFKDQVILL peptide in SARS-CoV-2, with the same binding pockets in positions 2, 4 and 9.
“These results suggest that both peptides can bind to HLA-A*2402 and provide targets for anti-viral T-cell responses, which are of great interest in light of the current pandemic,” said co-author Gregory Lizée, a professor in the Department of Melanoma Medical Oncology at MD Anderson. “But these results also shed light on many other potential immune targets, including those of other viruses and even human cancers.”
Kavraki noted that experimental work by longterm collaborator Lizée and Kyle Jackson, a graduate research assistant at Lizée’s lab who produced the mutant proteins, were critical to validate their simulations. Kavraki’s own lab won a National Science Foundation (NSF) Rapid Response Research grant to help identify fragments of SARS-CoV-2 viral proteins as possible targets for vaccine development.
Kavraki is the Noah Harding Professor of Computer Science and a professor of bioengineering, mechanical engineering and electrical and computer engineering.
The Cancer Prevention and Research Institute of Texas, the Gulf Coast Consortia, the NSF, the Einstein Foundation Berlin and the Welch Foundation supported the research.
Author: MIKE WILLIAMS